DUBAI: Researchers have identified a number of “silent” mutations in SARS-CoV-2, the virus behind the COVID-19 pandemic, a new study shows. The mutations were found within the roughly 30,000 letters of the virus's genetic code. The team of researchers, writing for the PeerJ journal, say that these minute changes helped the virus thrive once it made the leap from bats (and an intermediary wildlife) to humans – setting the stage for the global pandemic. Molecular biologists at Duke University said the subtle changes involved how the virus “folded” its RNA molecules within human cells.
30,000
letters that make up the SARS-CoV-2 genetic codeResults of the new study, published October 16, 2020, showed the statistical methods they developed to pinpoint the mutations — or “adaptive changes”— seen in the SARS-CoV-2 genome in humans [they did not do it on closely-related coronaviruses found in bats and pangolins]. "We're trying to figure out what made this virus so unique," stated lead author Alejandro Berrio, a postdoctoral associate in Greg Wray's lab at Duke University, in Durham, in the US state of North Carolina.
We're trying to figure out what made this virus so unique.
What’s different with this 'RNA folding' mutation, compared to previously seen mutations?
Previous research showed mutation markers within a gene that encodes the S "spike" proteins on the SARS-CoV-2 viral surface, which scientists explained play a key role in its ability to infect human cells, especially in the lungs. The new study also flagged mutations in the spike proteins. The confirmation of the S protein mutation meant that virus strains carrying these mutations were more likely to thrive.
Moreover, with the technique used by Berrio, Wray and Duke Ph.D. student Valerie Gartner, they helped identify new “culprits” – mutations inside the viral cover – that previous studies had failed to detect.
READ MORE
Moreover, with the technique used by Berrio, Wray and Duke Ph.D. student Valerie Gartner, they helped identify new “culprits” – mutations inside the viral cover – that previous studies had failed to detect.
What are the new 'silent' mutations seen?
There are at least two more: besides the "spike" protein (or exoprotein) mutations, the changes also occurred in two other regions of the SARS-CoV-2 genome – dubbed Nsp4 and Nsp16. These changes, they said, appear to have given the dominant virus strain an “edge” over previous strains, without altering the proteins they encode.
A number of studies have extensively investigated nonstructural proteins (Nsp), especially Nsp4 and Nsp16. For example, in 1998, the Journal of Virology published a study which showed evidence that mutations in rotavirus' Nsp4 are associated with altered virus virulence. More specifically, it showed that Nsp4 causes diarrhea in 6- to 10-day-old mice by functioning as an "enterotoxin" (i.e. a substance harmful to the digestive system).
In a 2015 study on SARS-CoV published in the Journal of Virology, the research team had homed in on nsp16 (nonstructural protein 16) and its role in facilitating viral replication and escaping innate immune recognition in host cells. The research team from China, included the legendary “Bat Woman” Zhengli Shi as well as Tero Ahola of Finland’s University of Helsinki. More importantly, researchers said the study made it possible to develop broad-spectrum "peptide inhibitors" by targeting the Nsp16 of coronaviruses.
Doing so, they explained, could strongly inhibit SARS-CoV viral replication and its disease-causing mechanism (pathogenesis). Another study done by Japanese researchers and published in 2017 (also in the Journal of Virology), pointed to the role of Nsp4, interacting with Nsp3, in boosting viral replication in SARS-CoV. Back then, scientists had already hypothesied that mechanisms underlying the interaction between these two Nsps (3 and 4), could guide the development of antivirals to treat coronavirus infections.
In the study published this October, Berrio's team took it further. They found that instead of affecting proteins (Spike), the new-found mutations likely affected how the virus's genetic material (made of RNA) “folds up” inside human cells. The researchers were quick to point out: they don’t know yet what sets these changes in the RNA structure in coronaviruses found in humans apart from coronaviruses in animals.
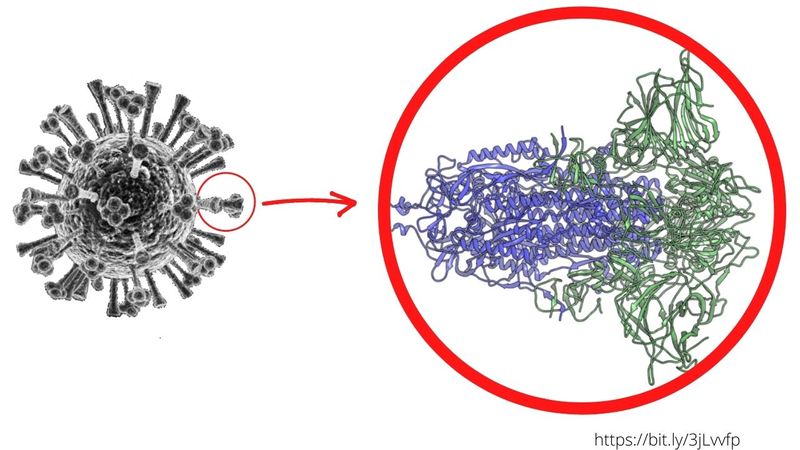
One thing is clear: these mutations have contributed to the virus's ability to “shed” efficiently – spreading the infection before people even know they have it. This particular property of SARS-CoV-2 (high viral “shedding” by carriers who are still “asymptomatic”) has made the current pandemic so much more difficult to deal with compared to the SARS coronavirus outbreak of 2003 (in which shedding took place after symptoms had manifested).
What’s the significance of these new-found mutations?
Researchers said their findings could lead to new targets for treating or preventing COVID-19 at the molecular level. "Nsp4 and Nsp16 are among the first RNA molecules that are produced when the virus infects a new person," Berrio explained. "The spike protein doesn't get expressed until later. So they could make a better therapeutic target because they appear earlier in the viral life cycle."
Nsp4 and Nsp16 are among the first RNA molecules that are produced when the virus infects a new person. The spike protein doesn't get expressed until later. So they could make a better therapeutic target because they appear earlier in the viral life cycle.
And by identifying the genetic changes that enabled the novel coronavirus to thrive among and transmit efficiently between human hosts, scientists hope to better predict future animals-to-humans disease outbreaks before they happen.
"Viruses are constantly mutating and evolving," Berrio told Science magazine. "So it's possible that a new strain of coronavirus capable of infecting other animals may come along that also has the potential to spread to people, like SARS-CoV-2 did. We'll need to be able to recognise it and make efforts to contain it early."
What is the RNA structure and function?
Unlike doubled-stranded helical structure of the DNA, the RNAs are single stranded, a linear polymer of ribonucleotides joined by 3-prime (or 3’) and 5-prime (or 5’) phosphodiester bonds. The RNA has four major bases: two Purine bases – Adenine [A] and Guanine [G]); and two Pyrimidine bases -- Cytosine [C] and Uracil [U].
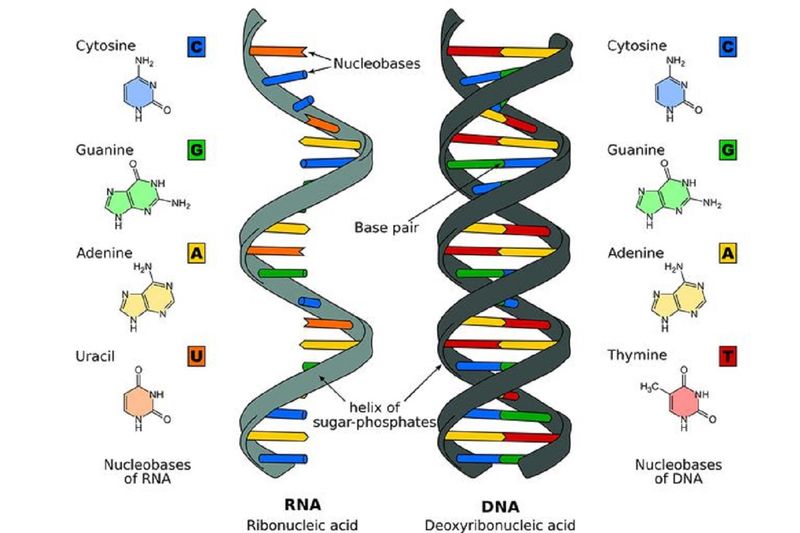
What is the difference between DNA and RNA?
There are two differences between DNA and RNA: (a) RNA contains the sugar ribose, while DNA contains the slightly different sugar deoxyribose (a type of ribose that lacks one oxygen atom); and (b) RNA has the nucleobase uracil while DNA contains thymine.
How do scientists observe RNA structure and 'folding'?
Using powerful electron microscopes with sub-nanometer resolution – and on millisecond timescales – scientists are able to observe the formation of biologically-active RNA structures. One technique involves the application of single-molecule fluorescence resonance energy transfer (sm-FRET). This technique reveal details of conformational changes and timescale of the process in RNA structure and “folding”. With further improvements, there emerged a more powerful tool to reveal information about the dynamics and energetics of RNA folding – which remains hidden using conventional sm-FRET approaches.
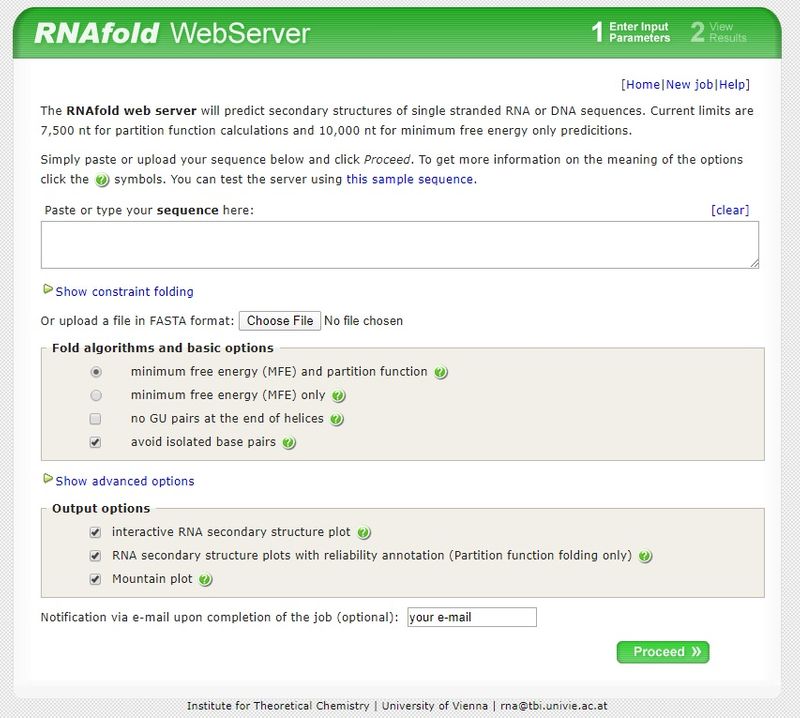
How do molecular biochemists study or predict RNA folding?
RNA secondary structure prediction is widely used to understand RNA function. To predict secondary structures of single-stranded RNA or DNA sequences, one tool scientists use is an online server, called the “RNAfold web server”. The server is run by the Institute for Theoretical Chemistry, at the University of Vienna in Austria.
from World,Europe,Asia,India,Pakistan,Philipines,Oceania,Americas,Africa Feed https://ift.tt/3knwzWv
No comments:
Post a Comment